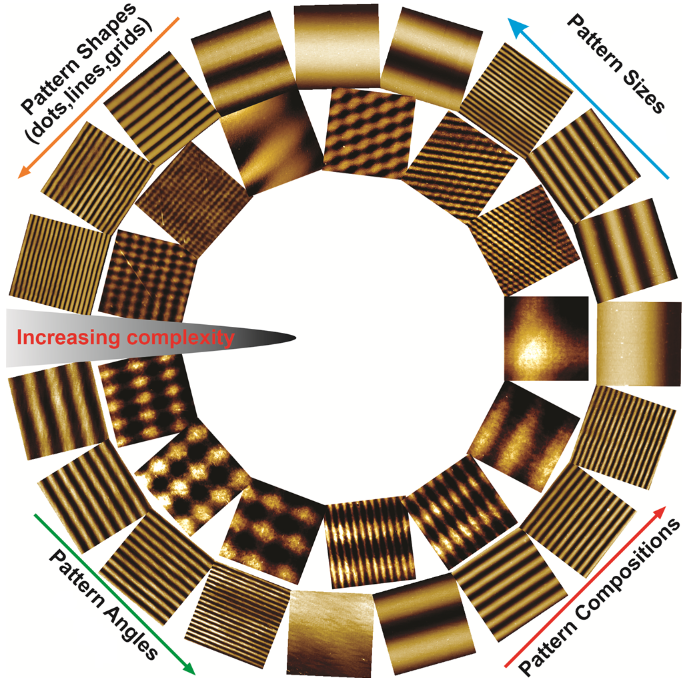
Scope of the combinatorial nanoarray cell screening platform – varying parameters can all be contained in the CBC array to match the complexity of natural ECMs and biophysical cues.
Invention Summary:
Biophysical cues mediated by natural or synthetic extracellular matrices (ECMs) play an essential role in cellular reprogramming. In nature, ECM is structurally complex, spanning a wide range of sizes and hierarchical topographies. However, biomaterials (e.g., hydrogels and nanofibers) used as a synthetic ECM for cellular reprogramming are often designed at a reduced complexity, leading to suboptimal cell-ECM models.
To address the aforementioned issues, Rutgers researchers (https://kblee.rutgers.edu/) have developed a Dynamic Interference Lithography (DIL) and applied the DIL to generate combinatorial ECM libraries with 104-106 unique structural elements for systematically investigating cell-ECM interactions. Further, a quantitative and predictive biophysical cue mapping and assessment approach has been developed to provide insight into ECM-directed cell behaviors such as proliferation, neuronal differentiation, and axonal growth by integrating machine learning-based analytics.
In short, the approach has been implemented and new ECM structures have been discovered for efficient reprogramming of fibroblasts into mature neurons within a short time frame.
Advantages:
- Capable of manufacturing large-scale, comprehensive, and indexable micro/nanoarrays in a highly cost-effective manner
- Highly scalable and reproducible
Market Applications:
- A method for manufacturing Combinatorial micro/nano arrays
- Diverse micro/nano arrays for screening unique nano-topographies that control specific cellular phenotypes
- A software for data analysis and nano-topography prediction
- Specific topography for converting skin cells into functional cells such as neurons
Intellectual Property & Development Status: Patent pending. Available for licensing and/or research collaboration.